Table of Contents
Update
I removed the images in the post, I hope the code is still useful to someone.
I'm not programming crystal anymore, maybe I'll add a new image at a later time.
For more information on this, see https://www.nature.com/articles/s41565-018-0337-2 .
Setup
If you want to follow along, you'll need to install crystal (or rewrite the code in some other language).
We need some way to read and write png images, so run
shards init
and
add following to your
shards.yml
file:
dependencies: stumpy_png: github: stumpycr/stumpy_png version: ~> 4.2.0
While reading the Handbook of Neuroevolution Through Erlang , I was surprise how simple the description of genetic algorithms (page 87) was:
- Create a seed population
- Create a fitness function
-
Loop until termination condition is reached
- Evaluate each organism's fitness
- Choose the fit organisms from the population
- Remove the unfit rest
- Create offspring from the fit ones
- Compose a new population from the offspring and their parents
Each organism needs a genotype (DNA), a phenotype (how it looks and behaves) and a function that map from one to the other.
I'll use color quantization, representing images with a limited number of colors, as an example. The genotype is a list of colors, the phenotype is how the input image looks after it has been reduced to these colors.
Organisms & Mapping
random_color
generates a random RGB color (duh) and
seed_random
creates a palette of
size
random colors.
require "stumpy_png" include StumpyPNG def random_color : RGBA RGBA.from_relative(rand, rand, rand, 1.0) end class Palette def initialize(@colors : Array(RGBA)) end def self.seed_random(size : Int32) : self new(Array.new(size) { random_color }) end # ...
quantize
maps from genotype to phenotype by finding the color in the
palette that is closest to the input color (has the smallest
distance
) for each
pixel in the image. To save some calculations later,
find_closest
returns the distance and the closest color. Using the squared distance
is faster and because
, it
won't affect the result.
Note: there are better ways to calculate the distance between two colors, this one is easy to implement good enough for now.
# ... # Squared euclidian distance between two colors def distance(a : RGBA, b : RGBA) : Int64 # Convert to Int64 first, otherwise the subtraction could underflow dr = a.r.to_i64 - b.r dg = a.g.to_i64 - b.g db = a.b.to_i64 - b.b dr * dr + dg * dg + db * db end def find_closest(input_color : RGBA) : {RGBA, Int64} best_color = RGBA::WHITE best_distance = Int64::MAX @colors.each do |color| distance = distance(input_color, color) if distance < best_distance best_color = color best_distance = distance end end {best_color, best_distance} end # ...
Now we can build the mapping function, the output / phenotype is a
Canvas
, a 2D grid of
RGBA
pixels, with quantized colors. Because
find_closest
returns a tuple, we need to use
[0]
to extract the
color.
# ... def quantize(image : Canvas) : Canvas Canvas.new(image.width, image.height) do |x, y| find_closest(image[x, y])[0] end end def write(image : Canvas, filename : String) StumpyPNG.write(quantize(image), filename) end # ...
The fitness function is pretty simple, we just iterate over all pixels
of the image and sum up the errors / distances. If one organism is
better
than another, the value
fitness()
is
smaller
.
# ... def fitness(image : Canvas) : Int64 image.pixels.reduce(0_i64) { |acc, col| acc + find_closest(col)[1] } end # ...
Lastly, we need some way to create offspring for the fittest organisms in our population.
There are various ways to do this:
- Mutation, copy the genes with a small chance that stuff gets messed up
- Crossover, combine the genes of multiple parents
- A combination of both
To keep the implementation as simple as possible, I'll only use the first one.
# ... def produce_offspring(chance) : self self.class.new(@colors.map { |color| mutate(color, chance) }) end def mutate(color, chance) rand < chance ? random_color : color end end
Evolutionary Loop
path
points to the image we want to quantize,
@population_size
tells
it how many organisms should be in each generation,
@colors
is the
length of each organism's palette / genes and
@mutation_chance
can be
used to adjust how probable mutations are.
@average_error
stores a history of fitness values for the whole
population, this is useful to see if the organisms actually improve.
@generations
keeps track on how many generations have passed, why this
is useful will become clear later.
class Quantizer @image : Canvas @population : Array(Palette) @population_size : Int32 @colors : Int32 @mutation_chance : Float64 def initialize(path, @population_size, @colors, @mutation_chance) @image = StumpyPNG.read(path) @average_error = [] of Float64 @population = Array.new(@population_size) { Palette.seed_random(@colors) } @generation = 0 end # ...
During each
step
of the evolutionary loop, we sort the organisms by
their fitness, store the average fitness in
@average_error
for later,
take the fittest half of the organisms and create a new population with
one offspring each.
evolve_to
just calls
step
until the target
generation is reached.
write_images
iterates over all organisms in the current population and
renders their phenotypes,
write_average_error
creates a
.csv
file
with the average error of each generation.
Math.log10
in combination with
.rjust(length, '0')
is just a trick
to prepend numbers with
0
so that they are all equally long
(e.g. =000=…=999= instead of
0
…=999=), this way image viewer
display them in the correct order.
# ... def score_fitness avg_error = 0.0 result = @population.sort_by do |palette| error = palette.fitness(@image) avg_error += error error end n = @population_size * @image.width * @image.height @average_error << Math.sqrt(avg_error / n) result end def step puts "Generation: #{@generation}" new_population = [] of Palette fittest = score_fitness[0...(@population_size / 2)] fittest.each do |r| new_population << r new_population << r.produce_offspring(@mutation_chance) end @population = new_population @generation += 1 end def evolve_to(generation) while @generation < generation step end end def write_images(path) Dir.mkdir_p(path) unless File.directory?(path) length = Math.log10(@population.size).ceil.to_i @population.each_with_index do |palette, i| name = path + i.to_s.rjust(length, '0') + ".png" palette.write(@image, name) end end def write_average_error(path) File.open(path, "w") do |file| @average_error.each_with_index do |error, gen| file.puts [gen, error].join(",") end end end end
Results
I'll use a qantizer with a population of 12 organisms, each with 20 colors and a 1% mutation chance as an example.
The input image is Lena, the standard test image .
q = Quantizer.new("./lena.png", 12, 20, 0.01) q.write_images("images/initial/") q.evolve_to(10) q.write_images("images/10/") q.evolve_to(100) q.write_images("images/100/") q.evolve_to(1000) q.write_images("images/1000/") q.write_average_error("avg_error.csv")
Initial (random seed population)
In the first (random) generation there already is one pretty good organism, the one at the top right.
After 10 generations it is the dominant version.
Finaly, after 1000 generations and lots of mutations, we arrive at something that looks surprisingly close to the input image.
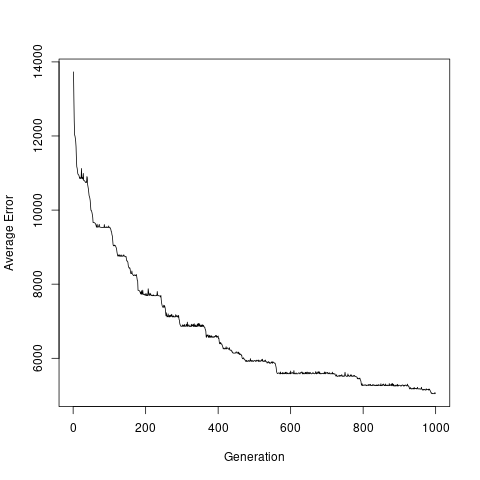
To confirm that the generations get better and better, we can plot the average errors collected along the way:
Images were created using the
montage
command that ships with
ImageMagick
.
Full source code: l3kn/genetic_color_quantization .